Summary
Stress responsive signalling coordinated by Nrf2 provides an adaptive response for protection of cells against toxic insults, oxidative stress and metabolic dysfunction. Nrf2 regulates a battery of protective genes by binding to regulatory antioxidant response elements (AREs). We found that Nrf2 is activated in cells without change in total cellular Nrf2 protein concentration. Regulation of ARE-linked protective gene transcription occurs rather through translocational oscillations of Nrf2. In live cell microscopy we observed that Nrf2 undergoes autonomous translocational frequency-modulated oscillations between cytoplasm and nucleus. Oscillations occurred in quiescence and when cells were stimulated at physiological levels of activators, they decrease in period and amplitude and then evoke a cytoprotective transcriptional response. We proposed a mechanism whereby oscillations are produced by negative feedback involving successive de-phosphorylation and phosphorylation steps. Nrf2 was inactivated in the nucleus and reactivated on return to the cytoplasm. Increased frequency of Nrf2 on return to the cytoplasm with increased reactivation or refresh-rate under stress conditions activated the transcriptional response mediating cytoprotective effects and links stress challenge to increased cytoplasmic surveillance. The serine/threonine-protein phosphatase PGAM5, member of the Nrf2 interactome, was a key regulatory component. We conclude that frequency modulated translocational oscillations of Nrf2 mediate the ARE-linked cytoprotective transcriptional response.
Background
Nuclear factor erythroid 2-related factor 2 (Nrf2) regulates the cellular expression of a battery of protective genes countering oxidative stress, toxic substances, lipid peroxidation, inflammation, metabolic dysfunction and ageing. It senses challenge to homeostasis in the cell cytoplasm and activates a protective transcriptional response. The human Nrf2 system regulates basal expression of ca. 640 genes, inducible expression of 650 genes and basal and inducible expression of 240 genes. Nrf2 activators typically affect ARE gene subsets; the selection mechanism is unknown but may be linked to the level of functionally active Nrf2 in the nucleus, accessory proteins (e.g. Maf proteins F, G & K) and pleiotropic effects of Nrf2 activators.
Under basal conditions, Nrf2 transcriptional activity is repressed by binding to Kelch-like erythroid cell-derived protein with CNC homology-associated protein 1 (Keap1). The Keap1-Nrf2 complex is in the cytoplasm and facilitates Nrf2 degradation: Keap1 is a substrate adaptor protein for Cullin-3 (Cul-3)-dependent E2 ubiquitin ligase complex, directing Nrf2 to the 26S proteasome. Keap1-Nrf2 binds threonine phosphatase PGAM5 tethered to the outer mitochondrial membrane.
Regulation of Nrf2
The mechanisms of stress sensing and surveillance by the Nrf2 system are uncertain. Two models have been proposed.
Stabilization model Under conditions of oxidative and other stresses Nrf2 is released from Keap1. An early mechanism of regulation of Nrf2 was stabilization for Nrf2 to proteolysis by release from KEAP1 and increased Nrf2 protein equilibration into the nucleus, binding to ARE sites for transcriptional regulation.
Oscillation model Nrf2 is phosphorylated by casein kinase-2 (CK2) and translocates to the nucleus via importins α5 and β1. In the nucleus, Nrf2 activates target genes by binding to AREs. Thereafter, Nrf2 is phosphorylated by Fyn kinase and expelled from the nucleus via the nuclear membrane export channel exportin-1/crm1. Nrf2 translocates to the nucleus driven by CK2 phosphorylation and activates the transcriptional response therein. Nrf2 is then inactivated by acetylation, expelled from the nucleus by fyn-catalysed phosphorylation and reactivated by deacetylase and phosphatase in the cytoplasm – see Figure 1. We presented an explanatory mechanism where Nrf2 functionality is linked to reactivation on return to the cytoplasm or “refresh rate”. Nrf2 is a constitutive oscillator where under conditions of oxidative and other stresses, the oscillation and refresh rate increase, increasing the level of active Nrf2 and thereby ARE-linked transcriptional response. The transcriptional response is linked to sensing of the cell stress status; increased stress increased stress status sampling frequency and also increases the transcriptional response. Nrf2 oscillations were found in the human endothelial cells and fibroblasts in culture.
Oscillation model – explanatory description
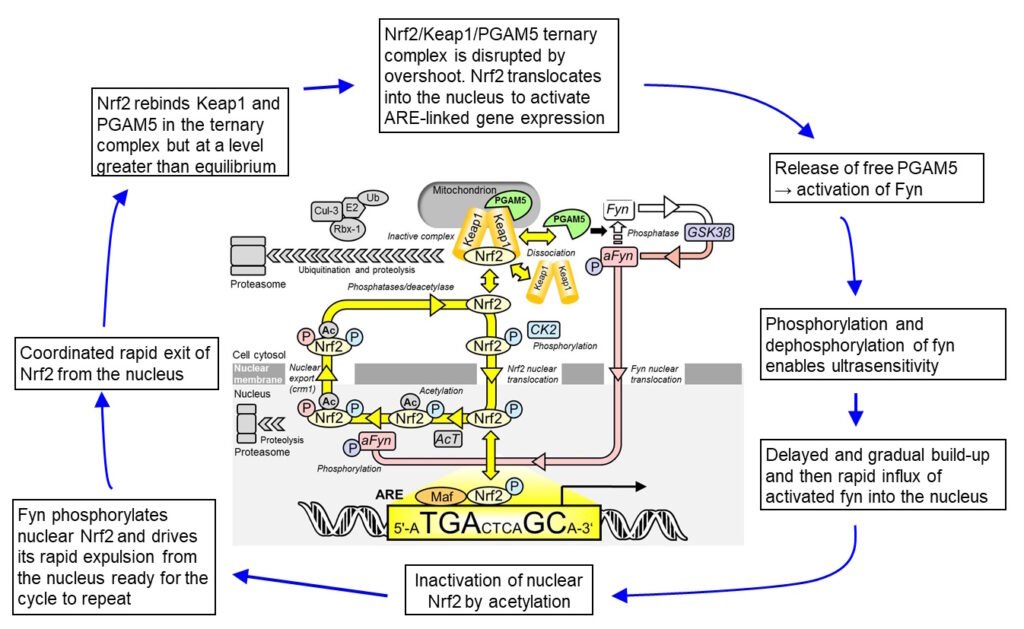
Figure 1. Oscillation mechanism for Nrf2 regulation. Abbreviations: a, activated; Ac, acetylation; AcT, acetyltransferase; CK2, casein kinase-2; Cul-3, Cullin-3; E2, ubiquitin E2 ligase; Fyn, fyn kinase; GSK3β, Glycogen synthase kinase-3 beta; Keap1, Kelch-like erythroid cell-derived protein with CNC homology-associated protein 1; Nrf2, nuclear factor erythroid 2-related factor 2; P, phosphorylation; PGAM5, mitochondrial serine/threonine protein phosphatase; Ub, ubiquitin.
Strengths of the oscillation model
Experimental observations explained by the Oscillation model but NOT by the Stabilization model
- Half-maximal transcriptional response is achieved without increase in Nrf2 protein
- Trapping of Nrf2 in the nucleus decreases transactivational activity
- Oscillation frequency responds to physiological levels or activators (10 – 20 μM hydrogen peroxide; 0.5 – 2 μM sulforaphane & 5 – 10 μM quercetin.
Oscillation model shows Nrf2 regulation is exquisitely fit-for-purpose in a stress-responsive transcriptional system
Because:
- Cytoplasmic stress sensing is communicated directly to the nucleus by Nrf2 translocation
- Frequency of stress surveillance is intensified when the host cell is challenged
- Inactivation of Nrf2 in the nucleus prevents abnormal accumulation of active Nrf2 and loss of transcriptional fidelity.
We noted analogy to an engineering sensor where dynamical resolution relies on a high refresh rate because the sensor must reassess the environment at a high rate. Nrf2 goes through cycles of sense> response> inactivate>reset. The nuclear translocation period of Nrf2 was 129 min in quiescent cells and decreased to 80 min during stimulus activation.
Oscillation model – experimental example
Time-lapsed video live-cell microscopy of human HMEC-1 endothelial cells transfected to express Nrf2-green fluorescence fusion protein and stimulated with 2 µM sulforaphane in vitro. Period of oscillation is ca. 80 min.
Oscillation model – molecular interpretation
Abbreviations are as given in Figure 1.
Oscillation model – transcriptional regulation
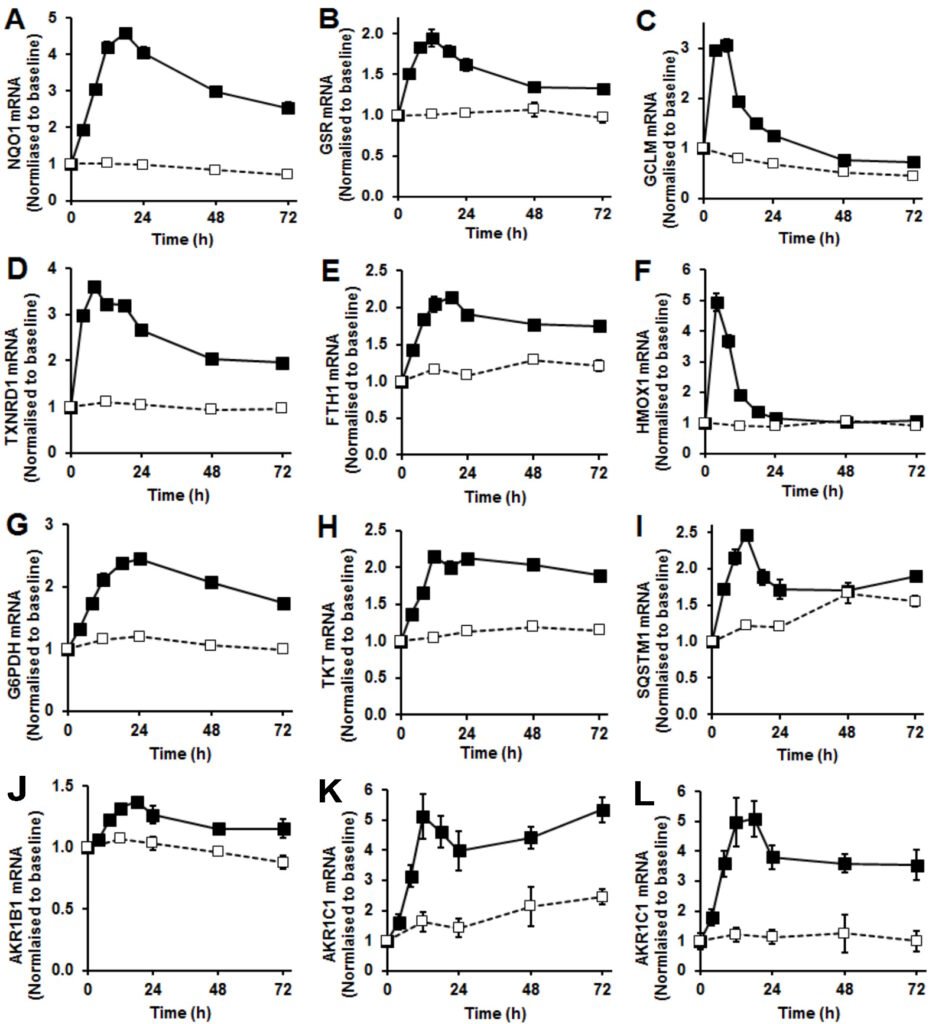
Figure 2 Nrf2 regulated gene expression. (A) – (L). Changes in gene expression, mRNA levels normalized to housekeeping genes in control (□ – – □) and sulforaphane-stimulated cells (■ – – ■). (A) quinone reductase NQO1, (B) glutathione reductase GSR, (C) γ-glutamylcysteine ligase (regulatory subunit) GCLM, (D) thioredoxin reductase TXNRD1, (E) ferritin FTH1, (F) heme oxygenase HMOX1, (G) glucose-6-phosphate dehydrogenase G6PDH, (H) transketolase TKT, (I) sequestosome-1/p62 SQSTM1 and (J), (K), and (L), aldoketo reductases isoforms 1B1, 1C1 and 1C3. Data are mean ± SD (n = 3), normalised to baseline level. Significance: all control and SFN time courses were significantly different in repeated measures analysis and at individual time points (P<0.001, t-test) except for HMOX1 from 24 – 72 h and SQSTM1 at 48 h. Indicators of significance are omitted for clarity.
Oscillation model – Sense>Response>Inactivate>reset cycle of the Nrf2 sensor
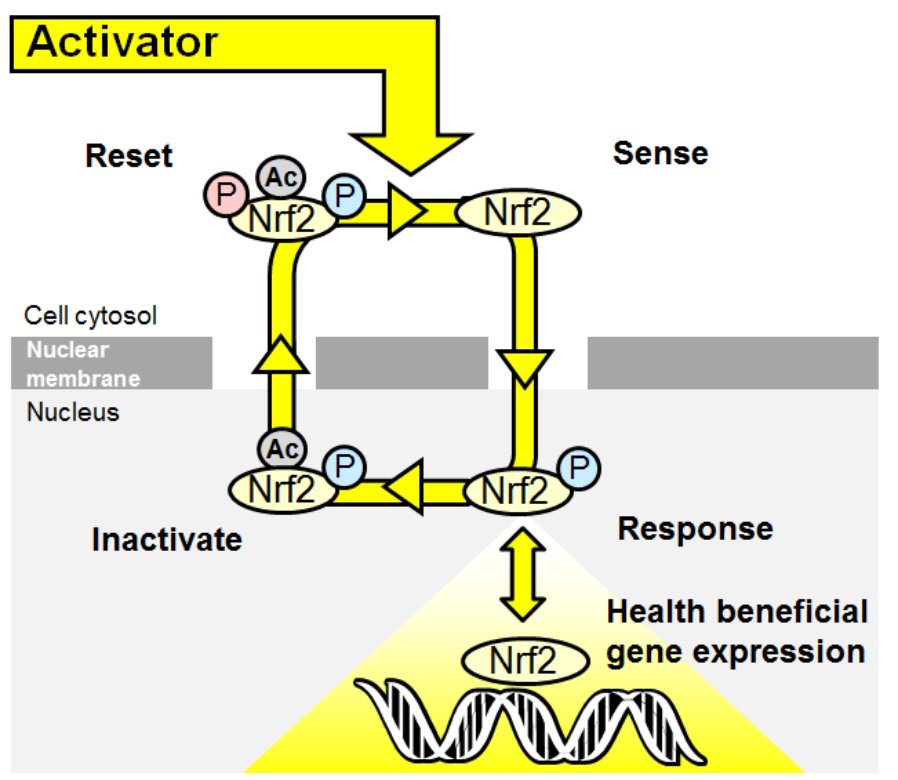
Figure 3. The Nrf2 transcriptional system as a wireless sensor.
Oscillation model – principal publications
- Xue, M., Momiji, H., Rabbani, N., Barker, G., Shmygol, A., Bretschneider, T., Rand, D.A. and Thornalley, P.J. (2015) Frequency Modulated Translocational Oscillations of Nrf2 Mediate the Antioxidant Response Element Cytoprotective Transcriptional Response. Antioxidants & Redox Signalling 23, 613 – 629.
- Xue, M., Momiji, H., Rabbani, N., Bretschneider, T., Rand, D.A. and Thornalley, P.J. (2015) Frequency modulated translocational oscillations of Nrf2 – a transcription factor functioning like a wireless sensor. Biochem. Soc. Trans.43, 669–673.
- Xue, M., Rabbani, N., Momiji, H., Imbasi, P., Anwar, M.M., Kitteringham, N., Parks, B.K., Souma, T., Moriguchi, T., Yamamoto, M. and Thornalley, P.J. (2012) Transcriptional control of glyoxalase 1 by Nrf2 provides a stress responsive defence against dicarbonyl glycation. Biochem. J. 443, 213 – 222.
- Xue, M., Weickert, M.O., Qureshi, S., Kandala, N.-B., Anwar, A., Waldron, M., Shafie, A., Messenger, D., Fowler, M., Jenkins, G., Rabbani, N. and Thornalley, P.J. (2016) Improved glycemic control and vascular function in overweight and obese subjects by glyoxalase 1 inducer formulation. Diabetes 65, 2282 – 2294.